Got Eclipse for PHP up running with XDebug
Thu, 2010-05-13 04:23 | by Samuel LampaGot Eclipse for PHP up running now, with XDebug. Yay :) It was a snap to install on my Ubuntu box. I basically followed this and this blog post. (The Ubuntu package for XDebug is php5-xdebug).
The Eclipse dialogs had changed location and structure a little, so for my documentation, I included a screenshot of the dialog under "Run > Debug configurations" below.

NSF soon requiring data management plans
Wed, 2010-05-12 19:24 | by Samuel LampaInteresting: "Scientists Seeking NSF Funding Will Soon Be Required to Submit Data Management Plans". Is this a new trend? If so, it should be an area where Bioclipse can help, I gues, not least through the planned ability to export semantic data to a Semantic MediaWiki (That will require me to finish my GSoC first, though).
Prolog query much faster when mimicking SPARQL
Sat, 2010-05-01 21:20 | by Samuel LampaI reported earlier that Jena/SPARQL outperformed Prolog for a lookup query with some numerical value comparison. It later on turned out that the results were flawed and finally that Prolog indeed was the fastest as soon as turning to datasets with more than a few hundred peaks.
The Prolog program I was using was rather complicated with recursive operations on double lists etc. Then, some week ago, I tried, in order to highlight differences in expressivity between Prolog and SPARQL, to implement a Prolog query that mimicked the structure of the SPARQL query I used, as close as possible. Interestingly it turned out that this Prolog query can be optimized to become blazing fast by reversing the order of shift values to search for, so that the largest values are searched for first. With this optimization the query outperforms both the SPARQL and the earlier used prolog code. See figure 1 below for results (The new prolog query is named "SWI-Prolog Minimal"). It appears that the querying time does not even increase with the number of triples in the RDF store!
 Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
The explanation seems to stem from the fact that larger NMR Shift values are in general more unique than smaller values (see histogram of shift values in the full data set in figure 2 below). Thus, by testing for the largest value first, the query will be much less prone to get stuck in false leads. (Well, looking at the histogram, it appears that one could in fact do even better sorting than just from larger to smaller, like testing for values around 100 before values around 130 etc.)
 Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
Automatically abbreviate authors first names in bibtex
Fri, 2010-04-30 17:12 | by Samuel LampaI had all my references (50+) with all author names spelled out, but was told to abbreviate first names to one letter (Indeed that looks better IMO too). I didn't want to do that manually, and found out that this can automatically be done by using the bibtex style "abbrv". So I set it in my latex document with
\bibliographystyle{abbrv} GSoC Project accepted
Tue, 2010-04-27 00:59 | by Samuel LampaJust got to know that my proposed GSoC 2010 project: "General RDF export/import in Semantic MediaWiki" (as documented here), was accepted! That's some good news! :). Mentor will be Denny Vrandečić from the Semantic MediaWiki community.
Surely the project is going to be a challenge but it is a highly motivating one so I'm much looking forward to it, to hopefully, together with my mentor and the community, to solve things, and to learn a lot.
I posted a (slightly shortened) copy of the project proposal and my bio here.
The project will be continuously documented here on this blog, so keep an eye here if you are interested (Use the GSoC2010 tag to filter out relevant posts). Community discussion will likely happen at the SMW-Devel mailing list, and if you want to contact me directly, you can do that at samuel dot lampa at gmail dot com or skype samuel_lampa.
My current status/schedule is:
- This week: Very busy, finishing thesis report.
- Next 2 weeks (though starting a little this week): Get dev. environment up running (leaning towards Eclipse with PDT) and looking at code
- 12/5: Briefing with Denny
- Up until 9th: Very busy period with exams on 24/5 and 8/6.
- On 9th: Start coding! (So coding start will be a little delayed, but will make up for that no worries! :) (not to used to having spare time anyway))
GSoC Proposal: "General RDF export/import in Semantic MediaWiki"
Tue, 2010-04-27 00:56 | by Samuel LampaThis is a slightly shortened version of the full Proposal, iniially posted on my user page on MediaWiki.org, and then in final form on the GSoC app site.
Browse semantic data in parallell
Mon, 2010-04-26 23:24 | by Samuel LampaThe semantic web field has seemed quite void of successful general user interfaces to browse semantic data in an efficient way (SPARQL querying is not really for everybody and their aunt). An interesting approach is the Freebase Parallax which lets you continuously views sets of data all while you narrow down or extend your search criteria, thus "browsing in parallell". Seems to make a lot of sence in its simplicity.
3rd Project Update (Integrating SWI-Prolog for Semantic Reasoning in Bioclipse)
Thu, 2010-04-08 14:17 | by Samuel LampaI just had my 3rd, and last project update presentation (before the final presenation on April 28th), presenting results from comparing the performance of the integrated SWI-Prolog against Jena and Pellet, for a spectrum similarity search query. Find the sldes below.
On larger datasets and more peaks in the query, Prolog is the fastest
Mon, 2010-03-29 16:21 | by Samuel LampaIn a previous performance test I compared a NMR Spectrum similarity search programmed in Prolog and run in SWI-Prolog integrated in Bioclipse, against a SPARQL Query doing the same task, run in Jena, and Pellet, also integrated in Bioclipse.
In that test Jena was the fastest, outperforming Prolog. The test had flaws though, as reported here with new tests added, though these new results were still not giving a clear winner.
This changed when I turned to larger datasets, testing on the range 2000-25000 spectra (where 25000 spectra corresponds to around a million RDF triples), instead of 10-100, which is a lot closer to the size of all spectra in NMRShiftDB (36419 searchable spectra as of March 29, 2010). I also used 16 instead fo 3 peak shift values in the search query. When testing with these changes, Prolog clearly took the lead.
(Bioclipse scripts, including the SPARQL Query, and the Prolog code, attached. See below).
This time I also included tests with Jena using both the in-memory RDF Store, and the Jena TDB disk based one. Interesting is that Jena run against the TDB disk based store is about double as fast as against the in-memory store! (I had to exclude the combination Pellet/TDB store, as it didn't complete for more than an hour, after which I stopped the run.)
To be noted, in these tests I did also take more measures as to avoid memory and/or caching related bias. For example, I did not rerun tests shortly after each other in a loop, but restarted Bioclipse after each run. This kind of testing took a lot more time, and I have so far only had time to do three iterataions for each specific size of dataset. Error bars, indicating the standard deviation, are included though to give an idea of the variation among the three. Hopefully I will have time to complement with a few more runs per measure point.
See figures below for results. I've included one figure where Pellet is included, and one where it is skipped, in order to get a zoom in on the Jena/Prolog difference.
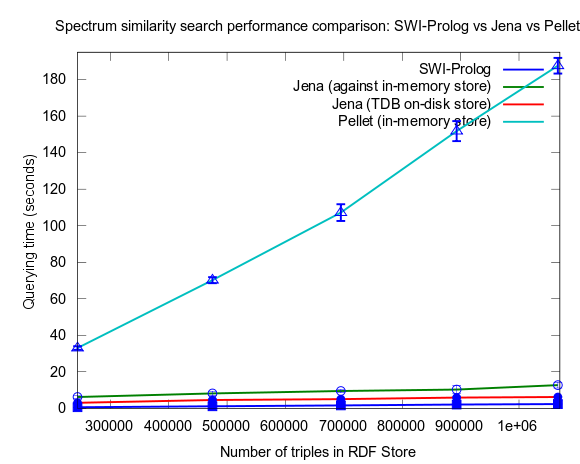
...and, Pellet excluded:
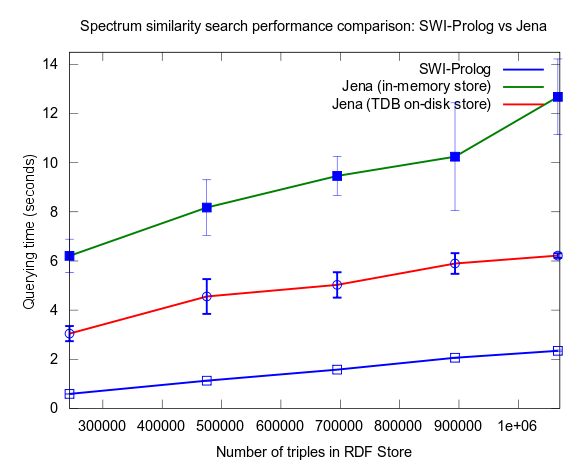
.png)