Pellet
3rd Project Update (Integrating SWI-Prolog for Semantic Reasoning in Bioclipse)
Thu, 2010-04-08 14:17 | by Samuel LampaI just had my 3rd, and last project update presentation (before the final presenation on April 28th), presenting results from comparing the performance of the integrated SWI-Prolog against Jena and Pellet, for a spectrum similarity search query. Find the sldes below.
On larger datasets and more peaks in the query, Prolog is the fastest
Mon, 2010-03-29 16:21 | by Samuel LampaIn a previous performance test I compared a NMR Spectrum similarity search programmed in Prolog and run in SWI-Prolog integrated in Bioclipse, against a SPARQL Query doing the same task, run in Jena, and Pellet, also integrated in Bioclipse.
In that test Jena was the fastest, outperforming Prolog. The test had flaws though, as reported here with new tests added, though these new results were still not giving a clear winner.
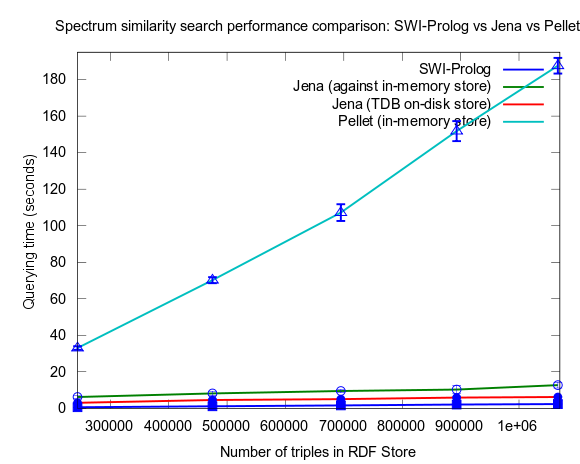
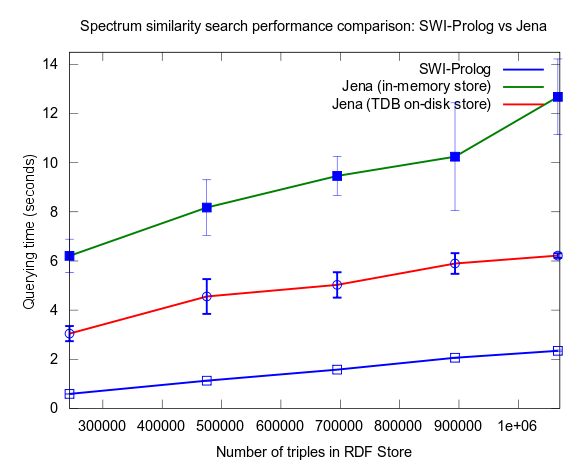
This changed when I turned to larger datasets, testing on the range 2000-25000 spectra (where 25000 spectra corresponds to around a million RDF triples), instead of 10-100, which is a lot closer to the size of all spectra in NMRShiftDB (36419 searchable spectra as of March 29, 2010). I also used 16 instead fo 3 peak shift values in the search query. When testing with these changes, Prolog clearly took the lead.
(Bioclipse scripts, including the SPARQL Query, and the Prolog code, attached. See below).
This time I also included tests with Jena using both the in-memory RDF Store, and the Jena TDB disk based one. Interesting is that Jena run against the TDB disk based store is about double as fast as against the in-memory store! (I had to exclude the combination Pellet/TDB store, as it didn't complete for more than an hour, after which I stopped the run.)
To be noted, in these tests I did also take more measures as to avoid memory and/or caching related bias. For example, I did not rerun tests shortly after each other in a loop, but restarted Bioclipse after each run. This kind of testing took a lot more time, and I have so far only had time to do three iterataions for each specific size of dataset. Error bars, indicating the standard deviation, are included though to give an idea of the variation among the three. Hopefully I will have time to complement with a few more runs per measure point.
See figures below for results. I've included one figure where Pellet is included, and one where it is skipped, in order to get a zoom in on the Jena/Prolog difference.
...and, Pellet excluded:
Correction of flawed results: Close competition between Jena and Prolog
Wed, 2010-03-24 04:50 | by Samuel LampaJena/SPARQL outperformed Prolog for spectrum similarity search
Thu, 2010-03-11 15:00 | by Samuel LampaUPDATE 29/3: See new results here
I was a bit worried over the performance of the RDF facilities in Bioclipse, as a SPARQL query for doing NMR Spectrum similarity search, including a numerical comparison run in Pellet (against datasets which are attached), were quite unsatisfactory, being some 2 orders of magnitude worse than some Prolog code I wrote for doing the same task (But of course, pure SPARQL with filtering is probably not what pellet is optimized for ...).
NMR SPARQL search problem solved
Fri, 2010-02-19 22:20 | by Samuel LampaThe problem I've had, to find a working SPARQL query for doing similarity search for spectra, according to a list of peak shift values (documented on this blog: post1, post 2 and on semanticoverflow) is now finally solved, thanks to helpful advice from Brandon Ibach on the [pellet-users] mailing list.
[solved] User defined datatypes not working in OWL 1.X
Mon, 2009-12-07 11:43 | by Samuel LampaI seemingly ran into the trouble that user-defined datatypes does not work in OWL 1.X (which is seemingly what the version of Pellet used in Bioclipse does support?
Performance comparison #2, Simple 13C Spectrum Similarity Search
Thu, 2009-12-03 15:53 | by Samuel LampaProlog
Bioclipse code
var start2 = new Date().getTime();
// js.say(blipkit.queryProlog( [ "findMoleculeWithPeakValuesNear", "100", "[23.3, 23.3, 23.5, 23.5, 26.1, 60.5, 90.0, 132.1, 0]", "Molecules" ] ));
js.say(blipkit.queryProlog( [ "findMoleculeWithPeakValuesNear", "100", "[12.5, 13.8, 23.8, 36.5, 44.3, 78.8, 87.3, 133.8, 0]", "Molecules" ] ));
var elapsed2 = (new Date().getTime() - start2)/1000;
js.say("Total time for finding molecule by shift values (Near-search): " + elapsed2 + " s");File based RDF storage in Pellet, first tests
Tue, 2009-11-24 17:32 | by Samuel LampaAs reported in a previous blog post, I ran into java stacksize errors when importing large amounts of data into pellet. Pellet was using just the in-memory Jena RDF store, which obviously puts limits on the amount of data it can handle.
Jena offers other options for RDF storage though, including SDB for SQL backends, and TDB for a pure Java file based storage.
Initial performance comparison: Pellet vs Prolog in Bioclipse
Mon, 2009-11-23 11:41 | by Samuel LampaI started with some initial performance testing for RDF data, between pellet an prolog, which are now both available integrated in Bioclipse.
Nice intro to RDF in Prolog (by Pellet author)
Wed, 2009-11-04 11:00 | by Samuel LampaI found a nice introduction to the use of RDF in Prolog (SWI-Prolog). It contains short primers for both RDF and Prolog, so it should be accessible to anyone with a minimal programming background: