Correction of flawed results: Close competition between Jena and Prolog
UPDATE 29/3: See new results here
I reported in a previous blog post (with a bit of surprise) that Jena clearly outperformed SWI-Prolog for a NMR Spectrum similarity search run inside Bioclipse. I have now realized that indeed these previous results were flawed for a number of reasons.
Firstly, the timing of the execution of the Prolog code included the loading of the RDF data into Prolog, which was not the case for Jena and Pellet. On the other hand, Jena and Pellet were running against a disk based RDF store, while Prolog was allowed to load the RDF data into memory, so the test was unfair both for Prolog and for Jena and Pellet.
I have now redone the tests, using instead the in memory RDF store of Pellet and Jena, and skipping the timing of RDF loading for Prolog. I also make sure to empty the variables that temporarily hold the in-memory stores, at each round. I also averaged over at least seven different runs for each tested reasoner.
Results
See figures below, and tabular data further below for results.
 SWI-Prolog vs Jena (NMR Spectrum Similarity Search)
SWI-Prolog vs Jena (NMR Spectrum Similarity Search)
The figure above shows that the comptetion between SWI-Prolog and Jena is indeed very tight for this number of spectra.
One puzzling thing is how SWI-Prolog performs better in unwarmed state than after running a few rounds(!). I suspect it could have to do with filling the memory stack with objects created by the Java Prolog Interface (JPL), which might not get the time needed to get cleared away by the garbage collector.
If that is the case, it could be a possible explanation for why SWI-Prolog in unwarmed state is increasing the execution time over the span of spectra (Prolog is in fact only completely unwarmed at the first run, while the others are run shortly afterwards in a loop)
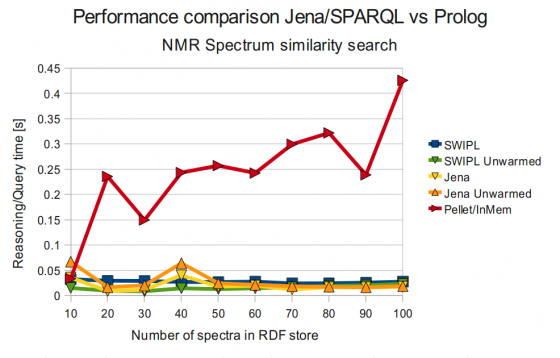 SWI-Prolog vs Jena vs Pellet (NMR Spectrum Similarity Search)
SWI-Prolog vs Jena vs Pellet (NMR Spectrum Similarity Search)
As can be seen in the figure above, Pellet was still performing quite poor, although far better than the earlier results!
Tabular data, SWI-Prolog vs Jena vs Pellet
No of spectra | SWIPL | SWIPL Unwarmed | Jena | Jena Unwarmed | Pellet |
| 10 | 0.032 | 0.015 | 0.036 | 0.067 | 0.034 |
| 20 | 0.029 | 0.010 | 0.009 | 0.017 | 0.236 |
| 30 | 0.029 | 0.009 | 0.011 | 0.020 | 0.150 |
| 40 | 0.027 | 0.014 | 0.040 | 0.064 | 0.243 |
| 50 | 0.027 | 0.013 | 0.018 | 0.024 | 0.257 |
| 60 | 0.028 | 0.014 | 0.020 | 0.021 | 0.242 |
| 70 | 0.024 | 0.017 | 0.012 | 0.018 | 0.300 |
| 80 | 0.024 | 0.019 | 0.017 | 0.017 | 0.322 |
| 90 | 0.026 | 0.020 | 0.016 | 0.016 | 0.239 |
| 100 | 0.028 | 0.023 | 0.017 | 0.019 | 0.426 |
Tests on larger RDF data sets now possible
The previously very poor results from Pellet was the reason why I choose to benchmark agains such small numbers of spectra (10 - 100) in the RDF store. Now that the Pellet reasoning doesn't take ages at least for few spectra, I might go on and do the test for some larger RDF data sets also.
I also realize there is some potential bias coming from running the reasoning over the different datasets closely after each other in a loop, since it might result in a buildup of objects in the memory stack, and also Prolog is not completely in an "unwarmed" state except for the first run (for 10 spectra).
Updated results coming soon.