SPARQL
My SMWCon Fall 2011 Talk now on YouTube
Mon, 2011-10-31 15:04 | by Samuel LampaI blogged it earlier, but better to get everything in one post, so taking the summary again:
After doing my GSoC project for Wikimedia foundation / Semantic MediaWiki in 2010, resulting in the RDFIO Extension, I finally could make it to the Semantic MediaWiki Conference, which was in Berlin in September.
Now, the video of my talk, "hooking up Semantic MediaWiki with external tools via SPARQL" (such as Bioclipse and R), is out on YouTube, so please find it below. For your convenience, you can find the slides below the video, as well as the relevant links to the different stuff shown (click "read more" to see it all on same page).
RDFIO 0.5.0 released
Fri, 2010-09-17 16:16 | by Samuel LampaVersion 0.5.0 of RDFIO, the MediaWIki extension providing PHP-based SPARQL endpoint and RDF import capabilities to to Semantic MediaWiki (and previously developed as part of my GSoC 2010 project ), is now released.
The 0.5.0 release
The 0.5.0 release fixes numerous bugs that were encountered as me and Egon Willighagen were working to hook up SMW with Bioclipse, as I blogged about earlier. We have now got this connection up running, so hopefully we tracked down most of the relevant bugs. A shortlist of changes can be found in the changelog. Links to download plus install instructions to be found on the extension page.
I hope to blog/screencast about the new Bioclipse->SMW editing functionality shortly.
RDFIO 0.3.0 released
Fri, 2010-07-30 04:54 | by Samuel LampaI just created a new release of the RDFIO MediaWiki extension. A somewhat detailed list of the changes can be found in the change log. The relevant links:
The filter by ontology / vocabulary feature
New for this release is a "export by ontology" feature, that - when possible - restricts the URIs used for a wiki page to only those that appears in an ontology that the user points to. To give an idea of this feature see the following screenshot:
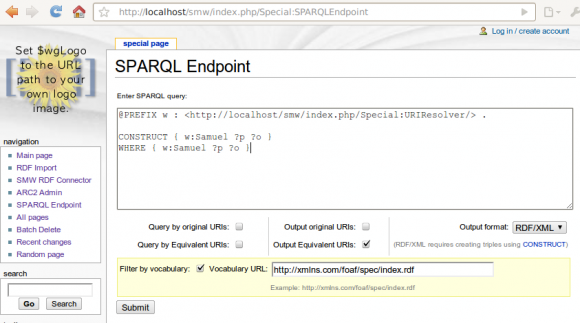
On the page "Samuel", I have one fact:
[[has blog::http://saml.rilspace.org]]
... and on the page "Property:has_blog", there are a number of facts, including:
[[Equivalent URI::http://xmlns.com/foaf/0.1/weblog]]
[[Equivalent URI::http://example.org/ExampleOntology/weblog]]
Current GSoC status
From the "remaining TODO list" from my last blog post, the following are finished with this release:
- In the SPARQL endpoint, enable querying by any URI specified as equivalent URI
- For RDF export, implement an "export by ontology" option, that - when possible - restricts the URIs used for a wiki page to only those that appears in an ontology that the user points to.
The remaining items ones are now:
- Create an HTML interface for interactively configureing how wiki titles should be chosen for RDF entities for which no preferred "wiki title property" (such as rdfs:label, dc:title etc.) was found.
- Add "pseudo namespaces" as an option for choosing wiki titles from general RDF URIs (not only properties!). I.e, the possibility to abbreviate a part of an URI into a pseudo-namespace, making the URI more fit for use as wiki title. (For properties, there if often a well known abbreviation for the corresponding vocabulary/ontology's base URI, but this is often not the case for general RDF entities, which can often be from some user defined data etc).
- If time permits:
- Implement filter by "export rdf" property.
Screencast: Installing Semantic MediaWiki and RDFIO from scratch on Ubuntu
Sat, 2010-07-17 01:48 | by Samuel LampaIn a previous blog post I demonstrated with a screen cast the RDFIO extension for Semantic MediaWiki but nothing on installation.
By testing I realized that the install procudure was VERY painful. I have now (with much valuable help from Oleg Simakoff) corrected a number of errors in the instructions and the code, and added to the install instructions commandline snippets for linux/ubuntu. I also created a screencast which goes through the steps from scratch (except Apache/MySQL/PHP setup), in a little more than 5 minutes. Hope this makes things easier for you testers! (And as you might try it out, please report any bugs or issues in the issue tracker!)
Sorry for the low volume level! Didn't realize that while recording ... :/
Screencast: RDF Import and SPARQL "Update" in Semantic MediaWiki
Fri, 2010-07-16 19:41 | by Samuel LampaSo, for those of you who might think the Install instructions for the RDFIO Semantic MediaWiki extension I'm working on are a bit daunting but would like a glimpse of what my GSoC project is up to anyway, I created a short (3:20) screencast demonstrating (ARC2 based) RDF Import and SPARQL "Update" functionality for some example data. (Sorry for the lame speaking ... :P ... didn't sleep for a looong time )
The screencast shows how you can import RDF/XML into Semantic MediaWiki and then use the SPARQL endpoint to insert or remove data to/from articles, even using the original format of the RDF that you imported earlier.
(For you who decide to try to install, please have a look at the error fixing happening in this thread.)
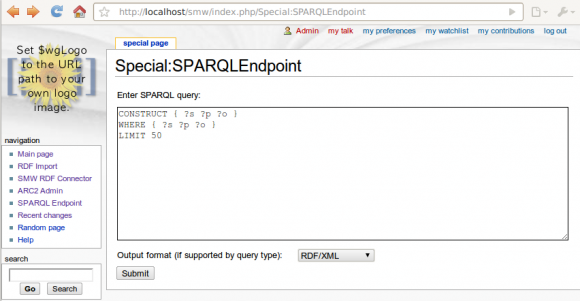
ARC2 based SPARQL Endpoint for Semantic MediaWiki up running
Tue, 2010-06-29 22:25 | by Samuel LampaI now managed to create a SPARQL endpoint for Semantic MediaWiki, in a MediaWiki SpecialPage, based on the ARC2 RDF library for PHP (including its built-in triplestore). See screenshot below, and code in the svn trunk. (I have to say I'm impressed by the ease of working with ARC2!)
Code is still quite ugly and the "Equivalent URI" handling that we talked about is still not implemented. Will turn to that now, while doing some refactoring (more object oriented etc).
Report approved - preliminary version for download
Thu, 2010-06-03 22:07 | by Samuel LampaMy degree project, titled "SWI-Prolog as a Semantic Web tool for semantic querying in Bioclipse" is getting closer to finish. Now my report is approved by the Scientific Reviewer (thanks, Prof. Mats Gustafsson), so I wanted to make it available here (Download PDF). Reports on typos are welcome of course! :)
Testing set up of SPARQL endpoint for SMW using RAP and NetAPI
Thu, 2010-05-13 04:25 | by Samuel Lampa(For my internal documentation, mostly)
Prolog query much faster when mimicking SPARQL
Sat, 2010-05-01 21:20 | by Samuel LampaI reported earlier that Jena/SPARQL outperformed Prolog for a lookup query with some numerical value comparison. It later on turned out that the results were flawed and finally that Prolog indeed was the fastest as soon as turning to datasets with more than a few hundred peaks.
The Prolog program I was using was rather complicated with recursive operations on double lists etc. Then, some week ago, I tried, in order to highlight differences in expressivity between Prolog and SPARQL, to implement a Prolog query that mimicked the structure of the SPARQL query I used, as close as possible. Interestingly it turned out that this Prolog query can be optimized to become blazing fast by reversing the order of shift values to search for, so that the largest values are searched for first. With this optimization the query outperforms both the SPARQL and the earlier used prolog code. See figure 1 below for results (The new prolog query is named "SWI-Prolog Minimal"). It appears that the querying time does not even increase with the number of triples in the RDF store!
 Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
The explanation seems to stem from the fact that larger NMR Shift values are in general more unique than smaller values (see histogram of shift values in the full data set in figure 2 below). Thus, by testing for the largest value first, the query will be much less prone to get stuck in false leads. (Well, looking at the histogram, it appears that one could in fact do even better sorting than just from larger to smaller, like testing for values around 100 before values around 130 etc.)
 Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
3rd Project Update (Integrating SWI-Prolog for Semantic Reasoning in Bioclipse)
Thu, 2010-04-08 14:17 | by Samuel LampaI just had my 3rd, and last project update presentation (before the final presenation on April 28th), presenting results from comparing the performance of the integrated SWI-Prolog against Jena and Pellet, for a spectrum similarity search query. Find the sldes below.