PROLOG
Final version of degree project report
Mon, 2010-07-05 21:01 | by Samuel LampaThe last administrative details of my thesis project are now finished, and the report is now available in final form, for download as PDF in on this page (no 14 in the list), or this direct link. (The title of the project was "SWI-Prolog as a Semantic Web Tool for semantic querying in Bioclipse: Integration and performance benchmarking").
Prolog query much faster when mimicking SPARQL
Sat, 2010-05-01 21:20 | by Samuel LampaI reported earlier that Jena/SPARQL outperformed Prolog for a lookup query with some numerical value comparison. It later on turned out that the results were flawed and finally that Prolog indeed was the fastest as soon as turning to datasets with more than a few hundred peaks.
The Prolog program I was using was rather complicated with recursive operations on double lists etc. Then, some week ago, I tried, in order to highlight differences in expressivity between Prolog and SPARQL, to implement a Prolog query that mimicked the structure of the SPARQL query I used, as close as possible. Interestingly it turned out that this Prolog query can be optimized to become blazing fast by reversing the order of shift values to search for, so that the largest values are searched for first. With this optimization the query outperforms both the SPARQL and the earlier used prolog code. See figure 1 below for results (The new prolog query is named "SWI-Prolog Minimal"). It appears that the querying time does not even increase with the number of triples in the RDF store!
 Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
Figure 1: Spectrum similarity search comparison: SWI-Prolog vs. Jena
The explanation seems to stem from the fact that larger NMR Shift values are in general more unique than smaller values (see histogram of shift values in the full data set in figure 2 below). Thus, by testing for the largest value first, the query will be much less prone to get stuck in false leads. (Well, looking at the histogram, it appears that one could in fact do even better sorting than just from larger to smaller, like testing for values around 100 before values around 130 etc.)
 Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
Figure 2: Histogram of NMR Shift values in 25000 spectrum dataset
3rd Project Update (Integrating SWI-Prolog for Semantic Reasoning in Bioclipse)
Thu, 2010-04-08 14:17 | by Samuel LampaI just had my 3rd, and last project update presentation (before the final presenation on April 28th), presenting results from comparing the performance of the integrated SWI-Prolog against Jena and Pellet, for a spectrum similarity search query. Find the sldes below.
On larger datasets and more peaks in the query, Prolog is the fastest
Mon, 2010-03-29 16:21 | by Samuel LampaIn a previous performance test I compared a NMR Spectrum similarity search programmed in Prolog and run in SWI-Prolog integrated in Bioclipse, against a SPARQL Query doing the same task, run in Jena, and Pellet, also integrated in Bioclipse.
In that test Jena was the fastest, outperforming Prolog. The test had flaws though, as reported here with new tests added, though these new results were still not giving a clear winner.
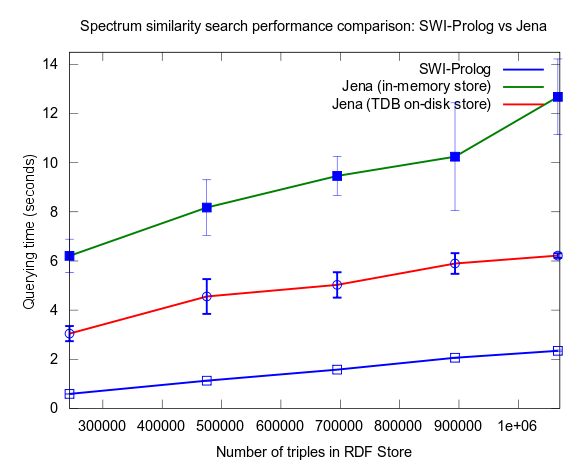
This changed when I turned to larger datasets, testing on the range 2000-25000 spectra (where 25000 spectra corresponds to around a million RDF triples), instead of 10-100, which is a lot closer to the size of all spectra in NMRShiftDB (36419 searchable spectra as of March 29, 2010). I also used 16 instead fo 3 peak shift values in the search query. When testing with these changes, Prolog clearly took the lead.
(Bioclipse scripts, including the SPARQL Query, and the Prolog code, attached. See below).
This time I also included tests with Jena using both the in-memory RDF Store, and the Jena TDB disk based one. Interesting is that Jena run against the TDB disk based store is about double as fast as against the in-memory store! (I had to exclude the combination Pellet/TDB store, as it didn't complete for more than an hour, after which I stopped the run.)
To be noted, in these tests I did also take more measures as to avoid memory and/or caching related bias. For example, I did not rerun tests shortly after each other in a loop, but restarted Bioclipse after each run. This kind of testing took a lot more time, and I have so far only had time to do three iterataions for each specific size of dataset. Error bars, indicating the standard deviation, are included though to give an idea of the variation among the three. Hopefully I will have time to complement with a few more runs per measure point.
See figures below for results. I've included one figure where Pellet is included, and one where it is skipped, in order to get a zoom in on the Jena/Prolog difference.
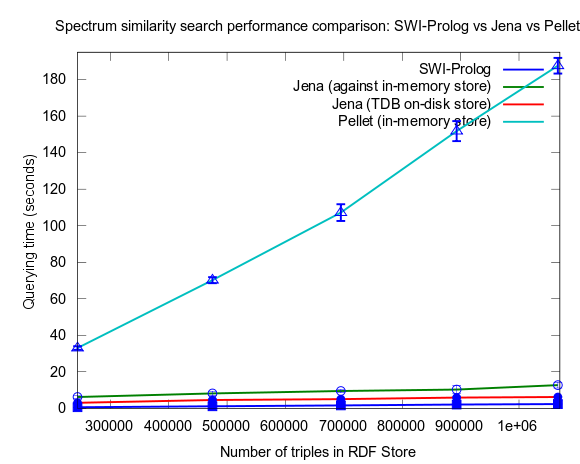
...and, Pellet excluded:
Need to add "colon dash" before rdf_register_ns
Fri, 2010-03-12 21:17 | by Samuel LampaI could not get the rdf_db:rdf_register_ns/2 function of SWI-Prolog's semweb package to work ... getting errors like "No permission to redefine static method ..." etc.
Now I finally figured out one has to add ":-" before the line with the rdf_db:rdf_register_ns predicate, like so:
:- rdf_db:rdf_register_ns(nmr, 'http://www.nmrshiftdb.org/onto#').
or ... inside Bioclipse:
swipl.loadPrologCode(":- rdf_db:rdf_register_ns(nmr, 'http://www.nmrshiftdb.org/onto#').");Spatial indexing for SWI-Prolog can be useful for modelling embryology with semantics
Fri, 2010-03-12 18:35 | by Samuel LampaJust found out that there is a Spatial Indexing package available for SWI-Prolog! How cool! That would come extremely handy if wanting to model the embryologic process of developmental biology with semantics (as I dream of doing :) ) ... and of course combining it with the ontologies and formats of the BioModels initiative.
Jena/SPARQL outperformed Prolog for spectrum similarity search
Thu, 2010-03-11 15:00 | by Samuel LampaUPDATE 29/3: See new results here
I was a bit worried over the performance of the RDF facilities in Bioclipse, as a SPARQL query for doing NMR Spectrum similarity search, including a numerical comparison run in Pellet (against datasets which are attached), were quite unsatisfactory, being some 2 orders of magnitude worse than some Prolog code I wrote for doing the same task (But of course, pure SPARQL with filtering is probably not what pellet is optimized for ...).
Screencast: Experimental Prolog integration in Bioclipse
Wed, 2010-03-03 20:51 | by Samuel LampaI wanted to test out some screen casting, so I chose to demo the (still experimental) SWI-Prolog integration into Bioclipse, showing how Prolog code (or a "Prolog knowledge base") can conveniently be stored inside Bioclipse's JavaScript environment (in a JS variable), loaded into the prolog engine, and then queried, all from the JS environment, and finally the results can be returned as well to the Javascript environment for further processing or output.
Note that this is still at the experimental stage, so things are a bit rough around the edges!
Chemical ring and cage structures need rules (seemingly)
Mon, 2010-03-01 23:31 | by Samuel LampaDuring my short stay at EBI, Egon had kindly arranged an opportunity to talk to Janna from the Steinbeck group, about some interesting work she has done on searching for cage structures in molecules, using Prolog, so we met over a coffee, together with Nico who's now at the Steinbeck group (visiting).
Bot Nico and Janna kindly gave a lot of good advice about research in general (as I'm currently looking into possibly doing PhD somewhere), which I highly appreciated. :)
And we talked some about the original topic too :), that is the cage structure problem. The cage structure problem is kind of an extension to the problem of expressing rings, which has previously been reported as a problem for OWL-DL. So because of this, it is interesting that Janna came up with a working solution, using Prolog.
As a highlighting example from the DL-side, Michel Dumontier has done some work on representing molecules, including rings. But they also had to use rules, not plain OWL.
So that seem to be the general conclusion: In order to express ring structures (or extensions of it, such as cage structures), you'll need to use rules in some way.
Unfortunatly my project is now running out of time, so I might not have much time to look more into this topic as part of my project :(. Will see if I can include this as a part of another course I still have to finish ("knowledge based systems in bioinformatics"), but that remains to see.
Think I finally got it what Horn clauses are good for
Thu, 2010-02-18 17:01 | by Samuel LampaThe Wikipedia article on Horn clauses states the following: "Horn clauses are relevant to theorem proving by first-order resolution, in that the resolution of two Horn clauses is itself a Horn clause"
That seems to explain why horn clauses are so foundational for prolog, since in prolog one can compose goal functions as compounds of other goals.
(I realize my lack of basic knowledge of Prolog, heavily regretting not having been able to have any formal training in logic programming / prolog at my uni :( (they removed the prolog part of the course where I hoped to learn it, just before I entered that course) ).