Samuel Lampa's blog
3rd Project Update (Integrating SWI-Prolog for Semantic Reasoning in Bioclipse)
Thu, 2010-04-08 14:17 | by Samuel LampaI just had my 3rd, and last project update presentation (before the final presenation on April 28th), presenting results from comparing the performance of the integrated SWI-Prolog against Jena and Pellet, for a spectrum similarity search query. Find the sldes below.
On larger datasets and more peaks in the query, Prolog is the fastest
Mon, 2010-03-29 16:21 | by Samuel LampaIn a previous performance test I compared a NMR Spectrum similarity search programmed in Prolog and run in SWI-Prolog integrated in Bioclipse, against a SPARQL Query doing the same task, run in Jena, and Pellet, also integrated in Bioclipse.
In that test Jena was the fastest, outperforming Prolog. The test had flaws though, as reported here with new tests added, though these new results were still not giving a clear winner.
This changed when I turned to larger datasets, testing on the range 2000-25000 spectra (where 25000 spectra corresponds to around a million RDF triples), instead of 10-100, which is a lot closer to the size of all spectra in NMRShiftDB (36419 searchable spectra as of March 29, 2010). I also used 16 instead fo 3 peak shift values in the search query. When testing with these changes, Prolog clearly took the lead.
(Bioclipse scripts, including the SPARQL Query, and the Prolog code, attached. See below).
This time I also included tests with Jena using both the in-memory RDF Store, and the Jena TDB disk based one. Interesting is that Jena run against the TDB disk based store is about double as fast as against the in-memory store! (I had to exclude the combination Pellet/TDB store, as it didn't complete for more than an hour, after which I stopped the run.)
To be noted, in these tests I did also take more measures as to avoid memory and/or caching related bias. For example, I did not rerun tests shortly after each other in a loop, but restarted Bioclipse after each run. This kind of testing took a lot more time, and I have so far only had time to do three iterataions for each specific size of dataset. Error bars, indicating the standard deviation, are included though to give an idea of the variation among the three. Hopefully I will have time to complement with a few more runs per measure point.
See figures below for results. I've included one figure where Pellet is included, and one where it is skipped, in order to get a zoom in on the Jena/Prolog difference.
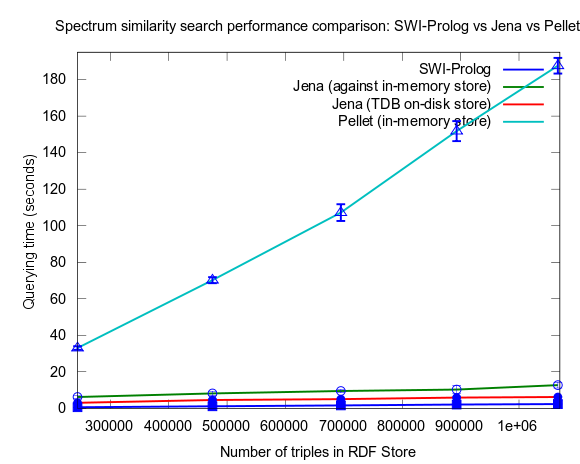
...and, Pellet excluded:
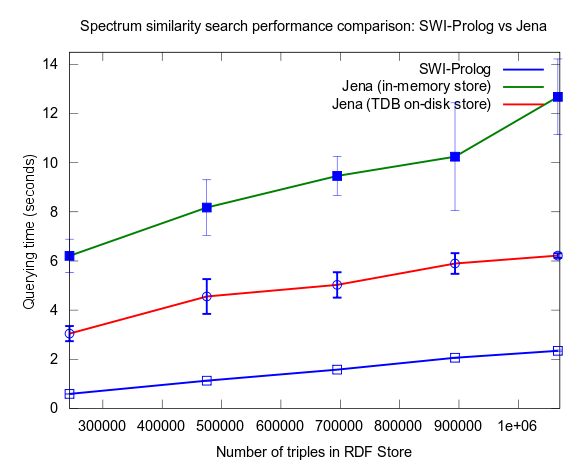
Correction of flawed results: Close competition between Jena and Prolog
Wed, 2010-03-24 04:50 | by Samuel LampaSemantic web for LAMP systems
Sat, 2010-03-20 14:45 | by Samuel LampaJust aquainted myself a tiny bit with ARC, RDF Classes for PHP. Might be useful in the Bioclipse / Semantic MediaWiki integration that is in the thoughts for the coming months. ARC "framework", or RDF API used by RDF modules in Drupal, and has been talked about being the substitute for the currently used RAP framework in Semantic MediaWiki (used if one wants to set up a SPARQL endpoint for a Semantic MediaWiki).
On a different note, there are a bunch of nice web apps built on ARC over at semsol.com, most notably trice, a semantic web application framework.
GSoC project suggestion
Fri, 2010-03-19 18:50 | by Samuel LampaIs there general enough import/export support for RDF data in Semantic MediaWiki that one could use SMW as a general collaborative RDF editor, as part of workflows?
(I know SMW has RDF export, but haven't yet found much about import (Except for OWL), and also wonder what is required to maintain the same RDF format "before and after" importing data for editing, and then exporting?)
I'm asking, firstly because we're interested in this functionality for integration with workflows made in Bioclipse's scripting environment (Since Bioclipse already has support for RDF storage, SPARQL querying,
Querying multiple SPARQL endpoints from single query, with Jena SERVICE extension
Tue, 2010-03-16 17:28 | by Samuel LampaEgon pointed to an interesting blog post about a feature that is available as a an extension to Jena, the semantic web framework available in Bioclipse. It allows to very easily query multiple SPARQL endpoints from a single SPARQL query (using the SERVICE keyword), and use variable bound from one endpoint when querying the next.
This is very useful in general. I was also thinking of the specific scenario (along the lines we have partly already been thinking) to use multiple Semantic MediaWikis as community maintained databanks, for querying back into Bioclipse. Being able to use multiple MediaWiki installs is very useful because it is hard to incorporate a very efficient access restriction system in MediaWiki (due to the nature of how it works, with template calls and all), so then it is better to be able to have separate wikis for content which needs special restrictions.
Need to add "colon dash" before rdf_register_ns
Fri, 2010-03-12 21:17 | by Samuel LampaI could not get the rdf_db:rdf_register_ns/2 function of SWI-Prolog's semweb package to work ... getting errors like "No permission to redefine static method ..." etc.
Now I finally figured out one has to add ":-" before the line with the rdf_db:rdf_register_ns predicate, like so:
:- rdf_db:rdf_register_ns(nmr, 'http://www.nmrshiftdb.org/onto#').
or ... inside Bioclipse:
swipl.loadPrologCode(":- rdf_db:rdf_register_ns(nmr, 'http://www.nmrshiftdb.org/onto#').");Spatial indexing for SWI-Prolog can be useful for modelling embryology with semantics
Fri, 2010-03-12 18:35 | by Samuel LampaJust found out that there is a Spatial Indexing package available for SWI-Prolog! How cool! That would come extremely handy if wanting to model the embryologic process of developmental biology with semantics (as I dream of doing :) ) ... and of course combining it with the ontologies and formats of the BioModels initiative.
Jena/SPARQL outperformed Prolog for spectrum similarity search
Thu, 2010-03-11 15:00 | by Samuel LampaUPDATE 29/3: See new results here
I was a bit worried over the performance of the RDF facilities in Bioclipse, as a SPARQL query for doing NMR Spectrum similarity search, including a numerical comparison run in Pellet (against datasets which are attached), were quite unsatisfactory, being some 2 orders of magnitude worse than some Prolog code I wrote for doing the same task (But of course, pure SPARQL with filtering is probably not what pellet is optimized for ...).
Getting the Blipstart files right
Wed, 2010-03-10 02:00 | by Samuel LampaI managed to fiddle a bit with my blipkit startup files, so I just need to document again
I use the files blipstart (a shell script), which loads the other file: blipstart.pl.
blipstart, the shell script, just contains the following line:
pl -L0 -G0 -A0 -T0 -q -g main -t hal -s /home/samuel/blipstart.pl
blipstart.pl is basically a copy of the blipkit/bin/blip file, removing the first line, and adding the following line at the end:
:- use_module(library('semweb/rdf_db')).