Samuel Lampa's blog
Eric Python IDE vs PyDev for Eclipse
Tue, 2010-12-28 15:54 | by Samuel Lampast discovered the Eric Python IDE, and I have to say I'm impressed.
I have been using PyDev for Eclipse so far, but was annoyed by the lack of options for the syntax highlighting, leaving me with rather sparingly colored code, which I found a bit hard to read at times. With Eric, I could configure up my favourite scheme (for which the cred goes to Rolf, my father :) ).
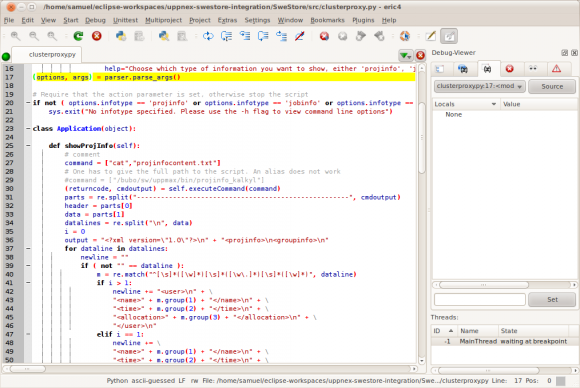
I have put the screenshots of my PyDev highlighting scheme, and the Eric one, below, so you can have a look for yourselves:
Eric Python IDE:
I will see soon which IDE I finally ended up using, but so far Eric seems to be the favourite ...
UPDATE: I have now found that Eric IDE lacks code navigation facilities (aka "Go to definition"), which makes it rather useless for my object oriented projects, where lots of code happens away in object methods...
UPPNEX web portal is live!
Wed, 2010-11-10 11:58 | by Samuel LampaUPPNEX's new web portal is now live!
- Visit: https://www.uppnex.uu.se
UPPNEX is a new initiative at Uppsala University, (lead by it's High Performance computing center SNIC-UPPMAX), to provide storage and high performance data analysis resources to the vibrant Next Generation Sequencing community in the Stockholm/Uppsala region and Sweden as a whole (some of the many recent projects was recently published in Nature).
This initiative is thought as a resource for wet-lab researchers with limited computer experience, and so it was important to provide with a one-stop place were these users can go to find documentation, information and contact to support staff. A website needed to be built.
Jonas Hagberg, system expert at UPPMAX and lead of the UPPNEX project, built up the site, created the current theme as a modification from the sky theme, and created the overall structure. I did - as currently working at Uppnex - come in at the later stage and created some graphics, the logo (in close collaboration with Jonas) and some additional configurations.
Swedberg lecture: Trying to survive the data deluge: bioinformatics tools for analyzing and visualizing large data samples
Mon, 2010-10-11 16:23 | by Samuel LampaDr. Reinhard Schneider from the European Molecular Biology Laboratory held a lecture at BMC in Uppsala with the title seen above. It seemed quite relevant to the stuff I'm currently doing at Science for Life Laboratory (where I'm employed for 2 months), investigating LIMS systems for NextGen sequencing data, as well as learning about analysis tools in the area.
What Reinhard presented was four different tools that they have developed/are working on, which tries to solve some of the problems of grasping heterogenous data sources. From the lecture info:
Editing Semantic MediaWiki from Bioclipse (with Screencast)
Fri, 2010-09-17 23:47 | by Samuel LampaThe original use case behind the RDFIO Semantic MediaWiki Extension which I developed as part of "Google Summer of Code 2010", and which was to hook up SMW with Bioclipse, is now concretizising. By using the new Bioclipse SMW Module (code here) it is now for the first time possible to add and remove SMW facts from inside Bioclipse, using a little Bioclipse JS Script:
var wikiURL = "http://drugmet.rilspace.org/wiki/"; smw.addTriple( "w:Caffeine", "w:is_a", "w:Molecule", wikiURL );
Removing triples is similar:
var wikiURL = "http://drugmet.rilspace.org/wiki/"; smw.removeTriple( "w:Caffeine", "w:is_a", "w:Molecule", wikiURL );
Well, you can use full URI:s also, but using the "w" prefix references wiki article titles directly. Thus you can view the result of the addition at http://drugmet.rilspace.org/wiki/Caffeine
What does this mean? Well, one thing is that with Bioclipse you can edit facts in SMW with the ease and power of javascript! This could enable scenarios where an SMW gets prepopulated with data for subsequent community editing, whereafter data can be transferred back to Bioclipse again, (as blogged about by Egon Willighagen already), possibly making community editing of scientific data mainstream!
There's a little convenience method for getting all RDF data from the SMW too:
rdfStore = rdf.createInMemoryStore(); rdfStore = smw.getRDF( "http://drugmet.rilspace.org/wiki/" );
... which you can then query locally with SPARQL, using the rdf manager:
result = rdf.sparql( rdfStore, "SELECT DISTINCT ?p WHERE { ?s ?p ?o } LIMIT 10" ) js.print( result );
...getting some output like so:
[["p"], ["http://www.w3.org/1999/02/22-rdf-syntax-ns#type"], ["http://www.w3.org/2000/01/rdf-schema#domain"], ["http://www.w3.org/2000/01/rdf-schema#range"], ["http://www.w3.org/2000/01/rdf-schema#subPropertyOf"], ["http://www.w3.org/2000/01/rdf-schema#subClassOf"], ["http://semantic-mediawiki.org/swivt/1.0#wikiPageModificationDate"], ["http://semantic-mediawiki.org/swivt/1.0#wikiNamespace"], ["http://www.w3.org/2000/01/rdf-schema#isDefinedBy"], ["http://semantic-mediawiki.org/swivt/1.0#page"], ["http://www.w3.org/2000/01/rdf-schema#label"] ]
That's it! Oh, well, I demonstrated the same thing with a screencast as well:
Yay! :) As getting this to work, we ran into a number of bugs in RDFIO, so that also resulted in a new release
RDFIO 0.5.0 released
Fri, 2010-09-17 16:16 | by Samuel LampaVersion 0.5.0 of RDFIO, the MediaWIki extension providing PHP-based SPARQL endpoint and RDF import capabilities to to Semantic MediaWiki (and previously developed as part of my GSoC 2010 project ), is now released.
The 0.5.0 release
The 0.5.0 release fixes numerous bugs that were encountered as me and Egon Willighagen were working to hook up SMW with Bioclipse, as I blogged about earlier. We have now got this connection up running, so hopefully we tracked down most of the relevant bugs. A shortlist of changes can be found in the changelog. Links to download plus install instructions to be found on the extension page.
I hope to blog/screencast about the new Bioclipse->SMW editing functionality shortly.
Autumn is here - new projects starting
Thu, 2010-09-09 17:03 | by Samuel LampaNow that the autumn is here I have some new projects starting, and running for approximately this month (before jumping out into the real world and trying to get a real job :) ). Good thing is, it all builds upon previous work.
First thing is, I'll work with Egon Willighagen to hook up Bioclipse with Semantic MediaWiki, via the RDFIO extension, which I developed as part of GSoC this year, mentored by Denny Vrandecic. I'm excited about putting RDFIO into some real world usage!
Second thing is I'll work part time at the Bioclipse group to improve the ways user documentation for plugins is authored and published, enabling some automation of publishing content to the Bioclipse website as well as the wiki etc. This will hopefully make it a lot easier for end users to find their desired extra functionality, and make more users see the value of a research platform with an open and modular structure, as is Bioclipse!
RDFIO 0.4.0 released - GSoC Finished!
Mon, 2010-08-16 18:55 | by Samuel LampaWith the release of RDFIO 0.4.0, my GSoC 2010 project is now over!
I want to thank especially my mentor Denny Vrandečić, and also the SMW community at large for a great time! I also want to sincerely thank my masters project mentor Egon Willighagen who mentioned about, and encouraged me to apply to the program. Without this encouragement, I'd never taken the step. It has been a good time and rewarding, and I've much enjoyed to have time to get a bit into MediaWiki/SMW extensions development, as well as to provide some new functionality to these great bits of software.
The main GSoC coding is now over, and I will need to take a little break for an exam this friday, but surely I'll continue to refine the RDFIO extension later, especially as Egon and me are looking into using it to integrate Bioclipse with SMW in order to make RDF data in Bioclipse "Community editable" (could turn out to be some real useful stuff!).
The 0.4.0 release
This new release brings a lot of refactoring and reworkings under the hood, as well as quite a few minor bugfixes and improvements here and there, so upgrading is recommended. We also got the issues in SMWWriter fixed now, so patching it is no longer needed, which hopefully will make installing easier!
- Se the extension page for more info about download and install
Of the more notable changes are the improved selection of wiki titles on import, as described in this blog post. Another important fix is that the default output format (if not specifying any) is now "SPARQL Resultset XML" which now makes the SPARQL endpoint fully "SPARQL compliant" and queryable from typical SPARQL tools like Jena. It is a remaining topic though how to allow update operations without leaving the endpoint wide open ... i.e. how to implement some form of user rights checking, when used as a webservice.
A little technical note also is that RDFIO now takes the $wgDBprefix parameter into account, so if you are using RDFIO with table prefixes in the database, you will need to regenerate the tables and the triples in the store (can be done at the Special:ARC2Admin and Special:SMWAdmin pages respectively).
I should not end without a note about some great bits of existing code that I've had the pleasure to make use of:
- ARC, the PHP RDF library I've been using, and on which RDFIO is very heavily dependent
I found it very powerful and super conveniant to work with! - I enjoyed making use of the SMWWriter and PageObjectModel extensions, which also definitely made life easier for me and saved me tons of work.
Sensible wiki titles on RDF import with "pseudo RDF namespaces"
Sun, 2010-08-08 02:35 | by Samuel LampaThis week I just finished the last remaining items on my todo list for my Google Summer of Code project, (which is available in the form of the RDFIO MediaWiki extension). Those things, which I also mentioned in my last blog post were to:
- Add ability to use ("pseudo") namespaces for general RDF entities (non-properties) in order to choose wiki titles for them on RDF import.
- Add a screen that shows URIs lacking a namespace prefix to abbreviate it with.
Regarding the first point, it might not be overly easy to see the usefulness of it at once, so I just created a screencast to show the difference between using it and not:
It demonstrates the problem of choosing sensible wiki titles for general RDF entities in case no good property for naming is available, (such as rdfs:label etc) ... since "entity" URIs often just consist of nonsensible id:s and often no namespace prefixes are defined for them. RDFIO lets you add "pseudo" namespaces (using a simplified splitting pattern, not necessarily consistent with XMLns specs), in order to come around this problem.
- The new functionality is so far only available in the svn trunk
- More info, install instructions etc on the extension page
Hopefully I'll find time to also demonstrate the second point above, as well as the "filter by ontology" feature for the SPARQL endpoint, with screencasts early next week.
Otherwise, the coming week I'll use for doing some refactoring of the currently quite unmanageable code, as well as add commenting, and hopefully also add the feature to filter RDF export by a [[Export RDF::false]] SMW property (which was the "it time permits" item of my TODO list).
RDFIO 0.3.0 released
Fri, 2010-07-30 04:54 | by Samuel LampaI just created a new release of the RDFIO MediaWiki extension. A somewhat detailed list of the changes can be found in the change log. The relevant links:
The filter by ontology / vocabulary feature
New for this release is a "export by ontology" feature, that - when possible - restricts the URIs used for a wiki page to only those that appears in an ontology that the user points to. To give an idea of this feature see the following screenshot:
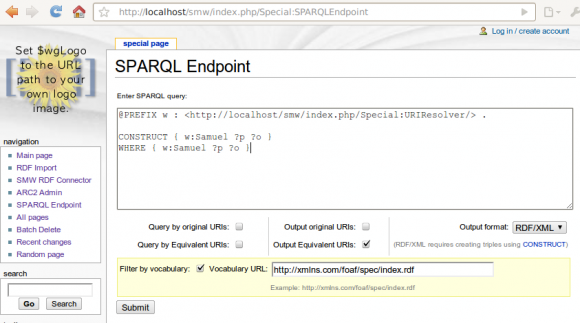
On the page "Samuel", I have one fact:
[[has blog::http://saml.rilspace.org]]
... and on the page "Property:has_blog", there are a number of facts, including:
[[Equivalent URI::http://xmlns.com/foaf/0.1/weblog]]
[[Equivalent URI::http://example.org/ExampleOntology/weblog]]
Current GSoC status
From the "remaining TODO list" from my last blog post, the following are finished with this release:
- In the SPARQL endpoint, enable querying by any URI specified as equivalent URI
- For RDF export, implement an "export by ontology" option, that - when possible - restricts the URIs used for a wiki page to only those that appears in an ontology that the user points to.
The remaining items ones are now:
- Create an HTML interface for interactively configureing how wiki titles should be chosen for RDF entities for which no preferred "wiki title property" (such as rdfs:label, dc:title etc.) was found.
- Add "pseudo namespaces" as an option for choosing wiki titles from general RDF URIs (not only properties!). I.e, the possibility to abbreviate a part of an URI into a pseudo-namespace, making the URI more fit for use as wiki title. (For properties, there if often a well known abbreviation for the corresponding vocabulary/ontology's base URI, but this is often not the case for general RDF entities, which can often be from some user defined data etc).
- If time permits:
- Implement filter by "export rdf" property.
New release of RDFIO (0.2.0) with security fixes
Thu, 2010-07-22 02:08 | by Samuel LampaJust to inform that I created a new release of the RDFIO MediaWiki extension. It contains important security fixes, by adding at least some basic checking of user rights and CSRFs (Cross site request forgeries) to the SPARQL endpoint, RDF import form etc. Thus, it's highly recommended to upgrade if you are using the extension on a public wiki!
- Download tarball
- Svn repo url: http://smwrdfio.googlecode.com/svn/branches/0.2.x
- Install instructions
Also, you might already have seen:
Current GSoC status
Otherwise, me and Denny just confirmed the remaining TODO list for my GSoC project, which is what I start working on now:
- In the SPARQL endpoint, enable querying by any URI specified as equivalent URI
- For RDF export, implement an "export by ontology" option, that - when possible - restricts the URIs used for a wiki page to only those that appears in an ontology that the user points to
- Create an HTML interface for interactively configureing how wiki titles should be chosen for RDF entities for which no preferred "wiki title property" (such as rdfs:label, dc:title etc.) was found.
- Add "pseudo namespaces" as an option for choosing wiki titles from general RDF URIs (not only properties!). I.e, the possibility to abbreviate a part of an URI into a pseudo-namespace, making the URI more fit for use as wiki title. (For properties, there if often a well known abbreviation for the corresponding vocabulary/ontology's base URI, but this is often not the case for general RDF entities, which can often be from some user defined data etc).
There are also some extra additions that I'll look into if time permits, like adding support for filtering the output on export with a property such as "RDF export::False", as suggested by Daniel Herzig.