Semantic MediaWiki
Working ARC2 RDF Store connector committed
Sat, 2010-06-19 02:28 | by Samuel LampaNow I have a working RDF Store connector for Semantic MediaWiki, that uses ARC2:s RDF store, rather than SMW:s built-in store. This will allow to take advantage of functionality in ARC2, such as possibility to set up a SPARQL endpoint etc.
Thanks to Alfredas Chmieliauskas for the Joseki store connector in the SparqlExtension for SMW, which this connector is heavily based upon.
The ARC2 connector implements the same amount of the SMWStore API as the JosekiStore, but I'm not yet sure if more needs to be implemented, for the things we want to do (general RDF import/export). Gotta figure that out.
The code is available in the google code repository trunk, and install instructions on the gcode wiki.
Feel free to try it out, but be warned that it has been only very briefly much tested at all yet!
Playing with SMWWriter
Thu, 2010-05-13 16:53 | by Samuel LampaWe are probably going to use SMWWriter for extending the RDF import/export functionality of Semantic MediaWiki, so I wanted to test it out a bit.
With some copy and paste of code from this page, I quickly had a MediaWiki Special Page set up, where I could make use SMWWriters internal API to implement a crude form for adding or removing "triples" in my Semantic MediaWiki. See Screenshot:
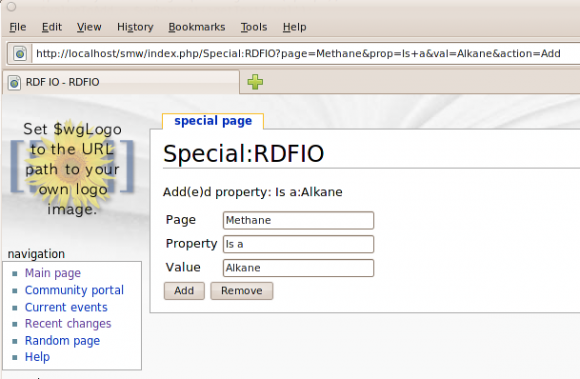
And the result, on the Methane page:
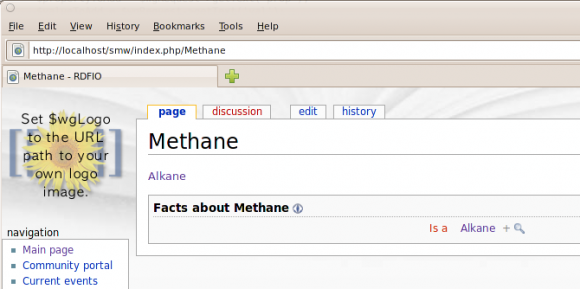
Looks promising. Connecting this with some ARC functionality for parsing SPARQL and RDF/XML, should make a big step in the right direction.
Testing set up of SPARQL endpoint for SMW using RAP and NetAPI
Thu, 2010-05-13 04:25 | by Samuel Lampa(For my internal documentation, mostly)
NSF soon requiring data management plans
Wed, 2010-05-12 19:24 | by Samuel LampaInteresting: "Scientists Seeking NSF Funding Will Soon Be Required to Submit Data Management Plans". Is this a new trend? If so, it should be an area where Bioclipse can help, I gues, not least through the planned ability to export semantic data to a Semantic MediaWiki (That will require me to finish my GSoC first, though).
GSoC Proposal: "General RDF export/import in Semantic MediaWiki"
Tue, 2010-04-27 00:56 | by Samuel LampaThis is a slightly shortened version of the full Proposal, iniially posted on my user page on MediaWiki.org, and then in final form on the GSoC app site.
Querying multiple SPARQL endpoints from single query, with Jena SERVICE extension
Tue, 2010-03-16 17:28 | by Samuel LampaEgon pointed to an interesting blog post about a feature that is available as a an extension to Jena, the semantic web framework available in Bioclipse. It allows to very easily query multiple SPARQL endpoints from a single SPARQL query (using the SERVICE keyword), and use variable bound from one endpoint when querying the next.
This is very useful in general. I was also thinking of the specific scenario (along the lines we have partly already been thinking) to use multiple Semantic MediaWikis as community maintained databanks, for querying back into Bioclipse. Being able to use multiple MediaWiki installs is very useful because it is hard to incorporate a very efficient access restriction system in MediaWiki (due to the nature of how it works, with template calls and all), so then it is better to be able to have separate wikis for content which needs special restrictions.
- « first
- ‹ previous
- 1
- 2
- 3