planet bioclipse
Systems biology simulation software for future Bioclipse integration?
Wed, 2010-02-10 02:52 | by Samuel LampaWhile reading up before the UU Cheminformatics journal club, initiated thanks to Egon Willighagen, I stumbled on this ... which seems to be something for Bioclipse, whenever it's time for extending it into the systems biology direction:
A LGPL licenced, Java based, Stochastic biological system simulator, designed with ease of integration and interoperability in mind. (Stochastical simulations seems to be the ones which currently captures biological behaviour the best).
Backtracking - Key difference between SPARQL and Prolog?
Tue, 2009-12-15 15:53 | by Samuel LampaOn something I realized a minute ago ...
Though being really different types of technologies, it might at first be tempting to compare a SPARQL query with a Prolog rule returning a list of results (Or at least it was to me until just a minute ago). In fact, SPARQL queries and prolog rules that return their results as lists, DO share some similarities. For example, in both you provide patterns of RDF statements with variables that are to be bound to each other or to RDF entities, in order to find all queried entities that match the pattern.
But then comes some important differences, in how SPARQL handles cases where one wants to evaluate looked-up entities with a function, like an arithmetic one. In SPARQL this has to be done (I think) with the FILTER construct, but that also means that backtracking is not done if nothing passes the filter (and that is the true meaning of a filter anyway, isn't it).
Bioclipse manager method to take arbitrary number of arguments
Tue, 2009-11-17 11:55 | by Samuel LampaI needed a Bioclipse manager method that could take an arbitrary number of arguments, (for a general purpose prolog method mapper). Through a useful discussion with jonalv, we figured out that there exists at least one working way of doing this, while there are a number of ways that do not work across both the Rhino/JavaScript though they work in Java alone.
A usage strategy emerges
Thu, 2009-11-12 17:31 | by Samuel LampaA strategy for how to work with the Bioclipse/JPL/Prolog/Blipkit combination I'm setting up, is becoming clear.
The main idea with Bioclipse, as well as with having a prolog engine available in it, is for flexible and "interactive" knitting together of knowledge. One of the main questions regarding how to use a Bioclipse/JPL/Prolog/Blipkit combination, has been where to put the bulk of knowledge integration/reasoning code? There would in principle be three options for that:
- Bioclipse (Javascript environment)
- The Blipkit-Prolog/Bioclipse integration plugin (Java code, a.k.a. "Manager methods")
- The prolog engine (As a prolog file)
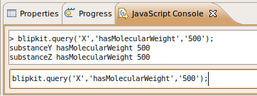
Testing out querying Prolog from Bioclipse
Sun, 2009-11-01 00:20 | by Samuel LampaMy project has been more or less on hold for around a week because of exams and other stuff. Looking forward to getting some concrete things done now. While still reading up a bit on OWL, I've started taken the first steps of the prolog/blipkit integration with a simple method for querying a prolog on the form "subject, a predicate, and an object".
Got blipkit started from inside Eclipse
Fri, 2009-10-16 14:55 | by Samuel LampaI now also managed to start blipkit from inside Eclipse.
The trick was to start the whole eclipse (The eclipse using for building Bioclipse) preceded with LD_PRELOAD=...the path to libjpl.so , and before that adding the paths to where libjava.so and libjvm.so are located.
SWI-Prolog JPL up running inside Bioclipse - Thinking about next steps
Wed, 2009-10-14 16:25 | by Samuel LampaSo, today I finally got the jpl (Java / Prolog API) up running inside Bioclipse, succeeding with compiling a simple test prolog program (The "test 0" in the "Test" example supplied with SWI-Prolog). Only talking to swi-prolog so far though, not blipkit. Starting blipkit includes loading prolog files etc. as specified in the blipkit startup script, so will have to study that in more detail.
"Refcards" for quickly getting into Bioclipse Plug-in development
Wed, 2009-10-14 16:00 | by Samuel LampaA link by Arvid led me to Dzone, where I found their "refcards": http://refcardz.dzone.com/ and after 15 minutes of reading, a lot of things, like file structure of Eclipse plug-in projects, had became much clearer.
For anyone new to Bioclipse plugin development, I think the following ones are the most relevant (especially the first two):
- Eclipse Plug-in Development
- Getting started with Equinox and OSGi
- Spring Configuration
- Spring Annotations
You will have to register at the site, and browse through the available ones in order to find them
Plugin development for Bioclipse - from scratch
Thu, 2009-10-08 10:51 | by Samuel LampaI switched laptop recently, so I needed to set up my development environment for Bioclipse 2.2 plugin development from scratch. Since the instructions for how to do this are spread over a couple of blogs and the wiki, I used the occasion to create an integrated howto, for my own documentation as well as for anyone interested.
Switching to Blipkit/BioProlog
Fri, 2009-09-18 14:24 | by Samuel LampaAfter installation problems with DR-PROLOG, and after looking closer to Blipkit/BioProlog (Biomedical Logic Programming Knowledge Integration Kit), after a suggestion by Claes Andersson, I now aim to integrate it instead of DR-PROLOG.